NumPyro Basics
Contents
NumPyro Basics¶
NumPyro is a probabilistic programming library that provides
Implementations of distributions, constraints and transforms
Primitives to specify elements in probabilistic models
Sampling (e.g. MCMC) and Optimization (e.g. VI) based inference algorithms
Numpyro uses jax as its backend for autograd and just-in-time (JIT) compilation to high performance hardware (GPUs/TPUs). NumPyro is very efficient but also lightweight and simple to use
See also
For more on Jax, see this NIPS 2019 talk by Skye Wanderman-Milne
Note
NumPyro re-implements another probabilistic programming library called Pyro. Their interface is very similar, but Pyro uses Torch instead of Jax as its backend
import holoviews as hv
hv.extension('bokeh')
import jax.numpy as jnp
import jax.random as random
import numpyro.distributions as dists
import numpyro
numpyro.set_platform("cpu")
numpyro.set_host_device_count(2)
numpyro.__version__
'0.9.1'
Distributions¶
Several distributions are implemented in the numpyro.distributions submodule
As an example let’s create a normal distribution. This requires specifying its mean though loc and its standard deviation through scale
z = dists.Normal(loc=0.0, scale=1.0)
z
<numpyro.distributions.continuous.Normal at 0x7f35d31b0370>
We can then retrieve the values of the parameters using
z.loc, z.scale
(0.0, 1.0)
PDF and CDF
Distribution objects have a method called log_prob to compute the logarithm of the probability density function
x = jnp.linspace(-5, 5, num=100)
log_pdf = z.log_prob(x)
hv.Curve((x, jnp.exp(log_pdf))).opts(width=500)
There is also the cdf method to compute the cumulative density function
cdf_x = z.cdf(x)
hv.Curve((x, cdf_x)).opts(width=500)
Sampling
To get random samples from a distribution we use the sample o rsample methods. The latter uses reparametrization to sample, for example in this case of the gaussian
is equivalent to the following reparametrization
This has several advantages that will be discussed in future lessons
Note
Not all distributions have rsample. The has_rsample attribute can be used to check if rsample is available
z.has_rsample
True
The sample and resample methods expects
pseudo random number generator key from
jax.randomthe shape of the sampled tensor:
sample_shape
We can use the latter to sample a matrix of normal samples from a scalar normal distribution
key = random.PRNGKey(1234)
z.rsample(key, sample_shape=(3, 3))
DeviceArray([[ 1.3284667 , 0.22779723, -0.5049753 ],
[ 1.3871989 , -0.7382383 , -0.56204265],
[ 0.5238776 , 0.29912323, -0.01665386]], dtype=float32)
Shape
We can obtain the dimensions of the distribution using the shape method
z.shape()
()
In this example it is an empty tuple because we created a scalar distribution
Consider the following normal distribution with vector parameters
z = dists.Normal(loc=jnp.array([0, 10]), scale=jnp.array([1, 1]))
z
<numpyro.distributions.continuous.Normal at 0x7f35d0728f10>
The shape in this case is
z.shape()
(2,)
Note
This corresponds to two independent Normal distributions, that can be sampled together for convenience
key = random.PRNGKey(1234)
z.rsample(key, sample_shape=(5, ))
DeviceArray([[ 1.3284667 , 10.2277975 ],
[-0.5049753 , 11.387199 ],
[ 0.56284374, 9.437958 ],
[ 0.5238776 , 10.299123 ],
[-0.01665386, 10.882737 ]], dtype=float32)
Primitives for probabilistic graphical models¶
Consider the following probabilistic model
This model can be represented using a graph as
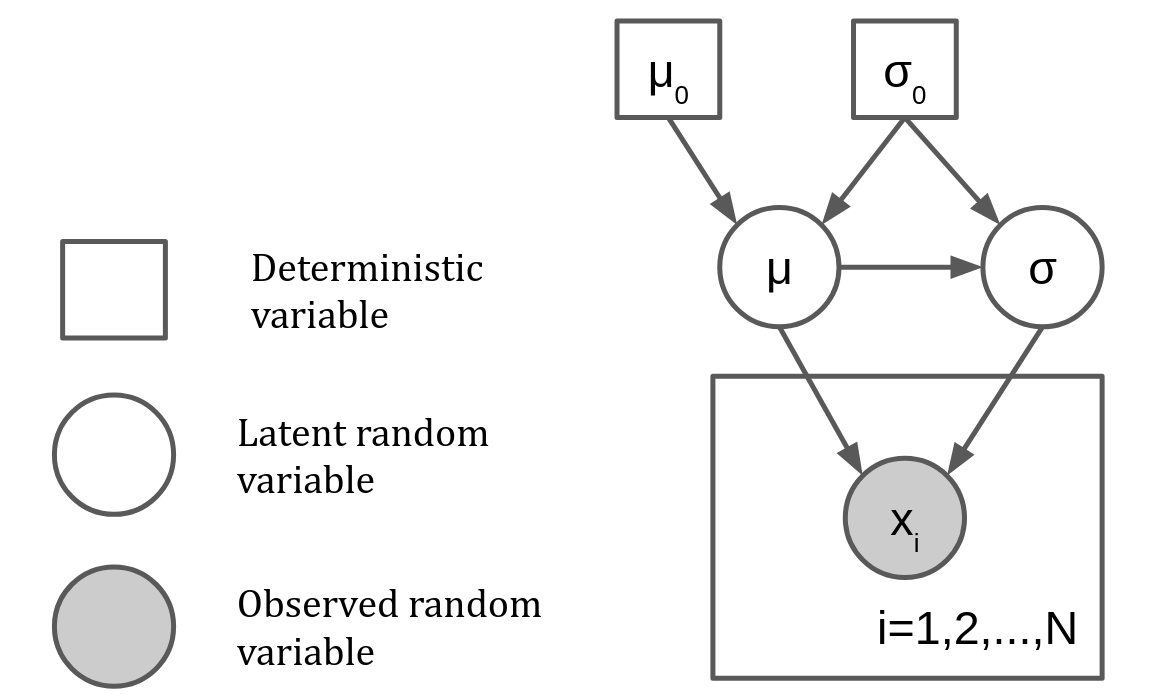
A probabilistic graphical model (PGM) is a statistical models were the relationships between variables (conditional dependence) are expressed through a graph. The key elements in PGMs are
Random variable (circle)
Latent: We don’t have samples from it
Observed: Wa have samples from it
Deterministic variable (square):
Hyperparameters with a fixed value
Result of deterministic function applied to random variables
Plate: Marks a group of variables that are conditionally independent given other variables
A PGM in NumPyro is a Python function that uses special primitives to represent these elements
Random Variables:
numpyro.sampleDeterministic variables:
numpyro.deterministicOptimizable deterministic variable:
numpyro.paramPlates:
numpyro.plate
In NumPyro jargon these are refered to as sites. Numpyro keeps track of all sites when inference algorithms are run with the model
The model mentioned before written with NumPyro primitives would be:
mu0, s0 = 0., 1.
def model(x=None):
mu = numpyro.sample(name='mu', fn=dists.Normal(loc=mu0, scale=s0))
s = numpyro.sample(name='s', fn=dists.LogNormal(loc=mu, scale=s0))
with numpyro.plate('data', size=len(x)):
numpyro.sample(name='x', fn=dists.Normal(loc=mu, scale=s), obs=x)
return mu, s # Return statement is optional
The sample primitive expects at least a unique name (
name) and a distribution (fn). We can also supply data using theobsargument to create an observed random variable. Otherwise we are creating a latent random variableThe plate primitive is used as a context and expects a unique name and a size. If the data is multidimensional
dimcan be used to set the axis of conditional indepedence
Note
Within a plate context manager, sample sites will be automatically broadcasted to the size of the plate
We will use the following synthetic data to learn our model
Warning
If we were to run model we would get an AssertionError. To sample from the model we have to specify a pseudo random seed
We can pass a seed to our model using the seed handler
x = dists.Normal(0., 5.).rsample(sample_shape=(20,), key=random.PRNGKey(0))
for seed in range(5):
with numpyro.handlers.seed(rng_seed=seed):
print(model(x))
(DeviceArray(-1.2515389, dtype=float32), DeviceArray(0.15910524, dtype=float32))
(DeviceArray(-1.1470195, dtype=float32), DeviceArray(0.10654547, dtype=float32))
(DeviceArray(-0.16715223, dtype=float32), DeviceArray(1.1439366, dtype=float32))
(DeviceArray(-0.05887505, dtype=float32), DeviceArray(2.5868776, dtype=float32))
(DeviceArray(0.78741604, dtype=float32), DeviceArray(1.2294954, dtype=float32))
Sampling based Inference: MCMC¶
With the model set we move on inferring the posterior of its parameters and predictions
First we will review Markov Chain Monte Carlo (MCMC) to perform sampling based inference. MCMC methods return samples from a markov chain that converges to the posterior we care. We will focus on the practical implementation of MCMC using numpyro.
See also
For more deep theoretical details on MCMC see Barber Chapter 27 or here
The main wrapper for MCMC methods in numpyro is
numpyro.infer.MCMC(sampler, # A sampler algoritm that decides the transitions
num_warmup, # Samples to discard in the beginning
num_samples, # Number of samples excluding the warmup ones
num_chains=1,
thinning=1, # Fraction of post-warmup samples retained
...
)
The main methods of MCMC are
run(): Populates the chain, expects the a PRNG key, the arguments of the model and initial values for the parametersget_sample(): Returns the posterior samplesprint_summary(): Returns a table with the statistics of the model parameters
Some of the samplers currently available are
Hamiltonian Monte Carlo:
HMCHamiltonian Monte Carlo for mixed discrete and continuous variables:
MixedHMCGradient-based Metropolis-Hastings:
BarkerMHNo-U turn:
NUTS
NUTS is a Hamiltonian methods that sets its step size automatically and is currently the state of the art. All kernels expects the function that specifies generative model plus their own particular arguments
%%time
from numpyro.infer import MCMC, NUTS
sampler = MCMC(sampler=NUTS(model, adapt_step_size=True),
num_chains=2, num_samples=1000, num_warmup=100, jit_model_args=True)
sampler.run(random.PRNGKey(1234), x)
/home/phuijse/.conda/envs/info320/lib/python3.10/site-packages/jax/_src/tree_util.py:185: FutureWarning: jax.tree_util.tree_multimap() is deprecated. Please use jax.tree_util.tree_map() instead as a drop-in replacement.
warnings.warn('jax.tree_util.tree_multimap() is deprecated. Please use jax.tree_util.tree_map() '
CPU times: user 9.71 s, sys: 43.2 ms, total: 9.75 s
Wall time: 9.81 s
Plot the statistical moments, \(\hat r\) statistic and 90% credibility interval
sampler.print_summary(prob=0.9)
mean std median 5.0% 95.0% n_eff r_hat
mu 0.98 0.61 0.97 -0.05 1.96 1698.90 1.00
s 6.02 0.98 5.88 4.60 7.53 1299.18 1.00
Number of divergences: 0
Get the samples and visualize the traces and posterior
samples = sampler.get_samples(group_by_chain=True)
def plot_trace(trace):
names = list(trace.keys())
plots = []
for name in names:
plot_param = []
for i, chain in enumerate(samples[name]):
plot_param.append(hv.Curve((chain), vdims=name, label=f'Chain {i}'))
plots.append(hv.Overlay(plot_param))
return hv.Layout(plots).cols(1).opts(hv.opts.Curve(width=600, height=150))
plot_trace(samples)
samples = sampler.get_samples(group_by_chain=False)
names = list(samples.keys())
joint = jnp.stack(list(samples.values())).T
distb = hv.Distribution(joint[:, 0], names[0]).opts(height=125) * hv.VLine(0.0) * hv.VLine(float(x.mean()))
distw = hv.Distribution(joint[:, 1], names[1]).opts(width=125) * hv.VLine(5.0) * hv.VLine(float(x.std()))
hv.Bivariate(joint, kdims=names).opts(cmap='Blues', line_width=0, filled=True, width=350) << distw << distb
Blue lines mark the true values used to generate the synthetic data
Red lines mark the MLE solutions